Gut Microbiota Composition and Functional Changes in Inflammatory Bowel Disease and Irritable Bowel Syndrome
By Dr. Katie E Golden, MD
The rapid evolution of microbiome research over the past decade has firmly established that our intestinal microbiota play an important role in a spectrum of human diseases. Inflammatory Bowel Disease, in particular, has repeatedly been associated with characteristic shifts in the bacterial populations that reside in the human gut. It has been a challenge, however, for researchers to better define the microbiome profiles and functional genomics that correlate with specific diseases. In a study published in Science Translational Medicine, the authors unveil a large-scale metagenomic microbiome analysis from patients with both Inflammatory Bowel Disease (IBD) and Irritable Bowel Syndrome (IBS).1 Their application of advanced biotechnology has identified key species and functions that distinguish these two disorders from each other and healthy controls.
IBD and IBS are gastrointestinal disorders that manifest in similar symptoms (abdominal pain, bloating, diarrhea and constipation), but with distinctively different pathophysiologies. IBD, which encompasses Crohn’s Disease (CD) and Ulcerative Colitis (UC), is characterized by chronic inflammation that is diagnosed by pathologic mucosal changes seen on intestinal biopsy. IBD, however, is poorly understood, there is no objective anatomic or pathophysiologic change, and therefore diagnosis is based on clinical symptoms. Microbiome research has been limited by low-resolution gene sequencing, and functional studies that only focus on specific bacterial species. In this study, researchers accelerated our investigatory approach by using shotgun metagenomic sequencing of nearly 1800 study subjects with IBD or IBS. Their findings further characterized bacterial growth profiles, antibiotic resistance, metabolic functions, and virulence factors to further explore the mechanisms that link microbiome changes to disease.
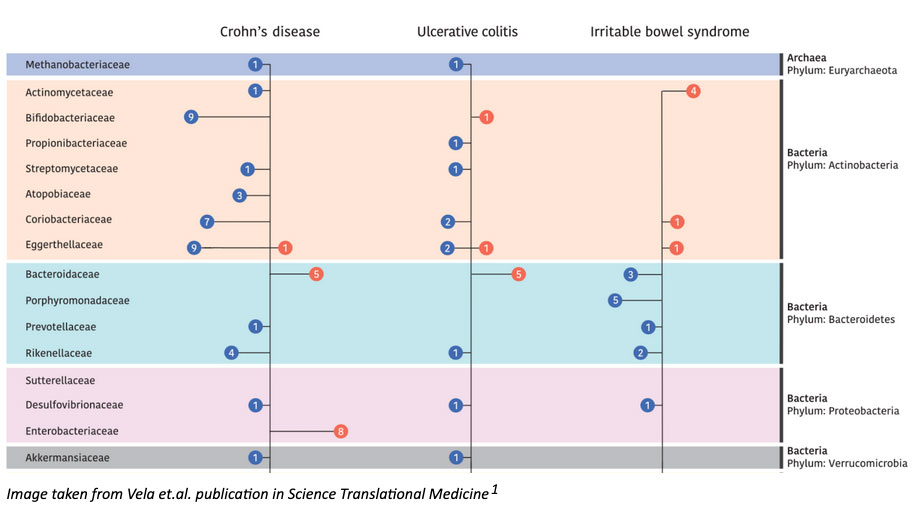
The authors discovered both overlapping and disease-specific associations when comparing microbiome profiles of IBD and IBS. To be more specific, they identified 24 taxa that were associated with both conditions (and different from control signatures), as well as a consistent increase in pathogenic strain diversity and reduced beneficial strain diversity in disease. They also observed that certain species were specific to each disease process (such as Bacteroides to IBD, and Streptococcus to IBS). To better understand the dynamic changes of microbial ecosystems, researchers also analyzed changes in bacterial growth rates, functional changes of the microbiome, changes in virulence factor production, and antibiotic resistance in disease-associated strains. These subsequent investigations led to a number of relevant findings. They observed reductions in methagenosis and hydrogen sulfide- producing pathways in IBD, alterations in microbial iron uptake pathways and increased MU-toxins in CD, and increased antibiotic resistance genes in both IBD and IBS compared to controls. While these are just a few examples, their expansive analysis provides several new opportunities for therapeutic development that respects the range of phenotypes in these heterogeneous diseases.
1. Vich Vila, A. et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci. Transl Med. 10, eaap8914 (2018).