Paper-Based Diagnostics for IBD
Primary Researchers:
Principal Investigator: James Collins, MIT
Project Summary:
Many new therapies for inflammatory bowel disease (IBD) have been developed, among which are biologics, a class of medications that function by reducing inflammation. The members of this medication class function by inhibiting different inflammatory pathways. Two major types of biologic medications are anti-tumor necrosis factor agents (anti-TNFs) such as infliximab [Remicade] or adalimumab [Humira] and anti-integrin agents such as vedolizumab [Entyvio]. Biologic medications are highly effective but only work for a subset of patients, and there are currently no standard methods to determine which patients will respond to which specific medications. These drugs are very expensive and often take multiple doses over months to achieve levels in the body that are needed for optimum function. The current approach to selecting these medications is based on trial and error, which leads to prolonged patient suffering and increased cost of care.
Recent studies have shown that changes in the gut microbial communities and in the function of gut cells in IBD patients may predict future response to two major categories of biologic medications, anti-TNF and anti-integrin agents. The results from these preliminary studies are promising, but their clinical application has been limited by the lack of an easy-to-use and affordable way to assess the gut microbial communities in patients. Standard methods to analyze these microbial communities are based on amplification and sequence identification of nucleic acids isolated from patient stool samples. These techniques are labor-intensive, time-consuming, cost nearly $1,000 per sample, and require high levels of technical training for analysis.
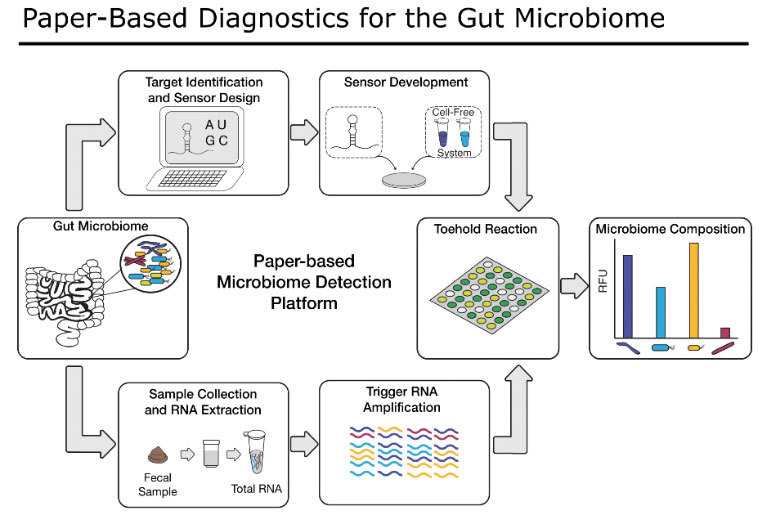
Here, we are adapting our previously-developed, highly innovative paper-based synthetic biology platform to assess members of the human gut microbial community, as well as human gut cell functions that are important for predicting response to IBD medications. Our technique costs pennies per reaction, is capable of detecting and quantifying nearly any nucleic acid target, and is simple to use. If successful, our sensors will improve the lives of IBD patients by providing an easy and accessible test for providers to help select the optimal medical treatment for each patient. Our flexible platform will also be able to incorporate new diagnostic targets from ongoing research by the IBD scientific community and continue to evolve as new therapies as associated tests are developed.